|
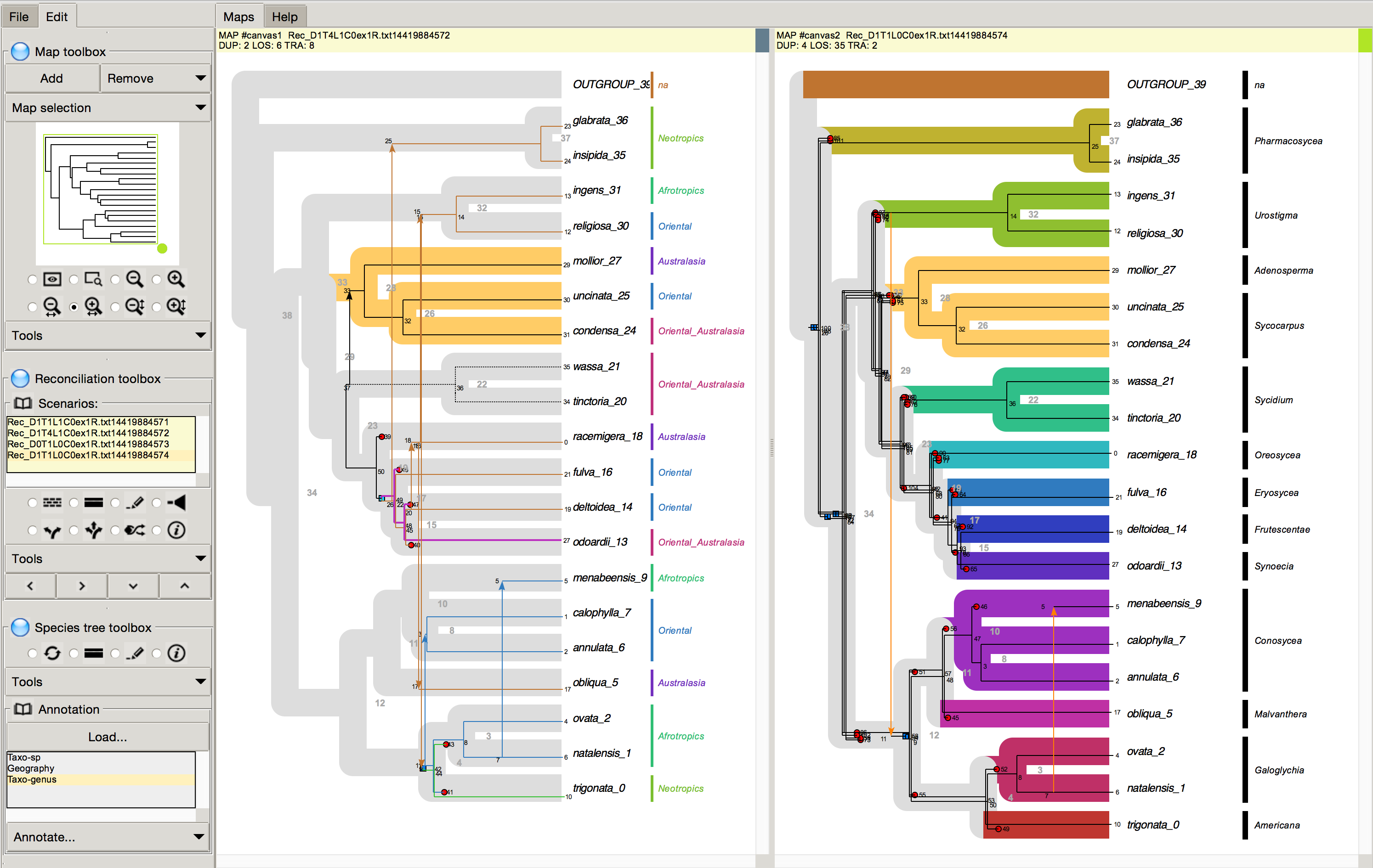
 Coming soon ! SylvX compatible with RecPhyloXML - a format for reconciled gene trees
Coming soon ! SylvX compatible with RecPhyloXML - a format for reconciled gene trees
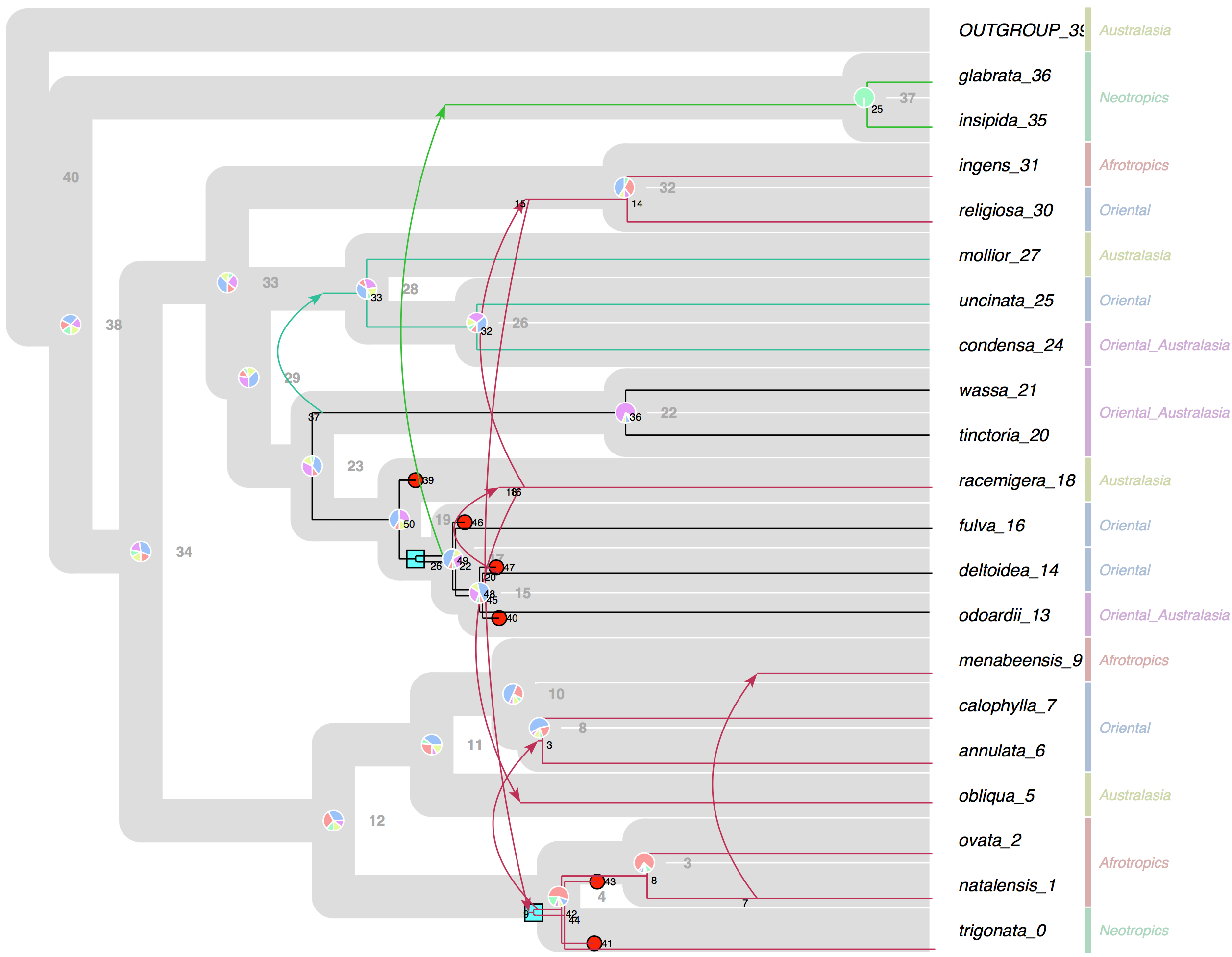
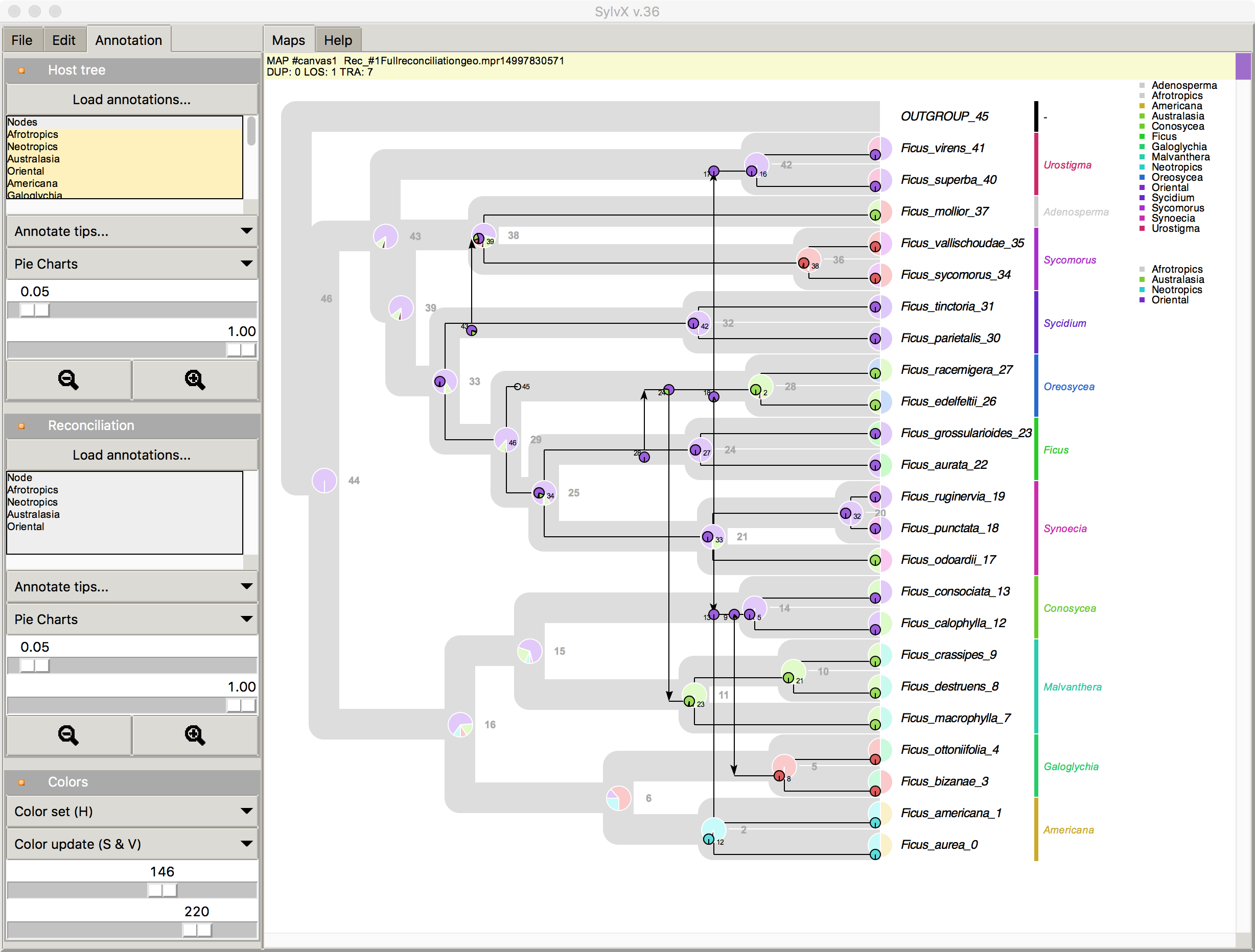
 Jul 2017 SylvX v.36 Annotation of Host tree and Reconcilaition
screenshot
Jul 2017 SylvX v.36 Annotation of Host tree and Reconcilaition
screenshot
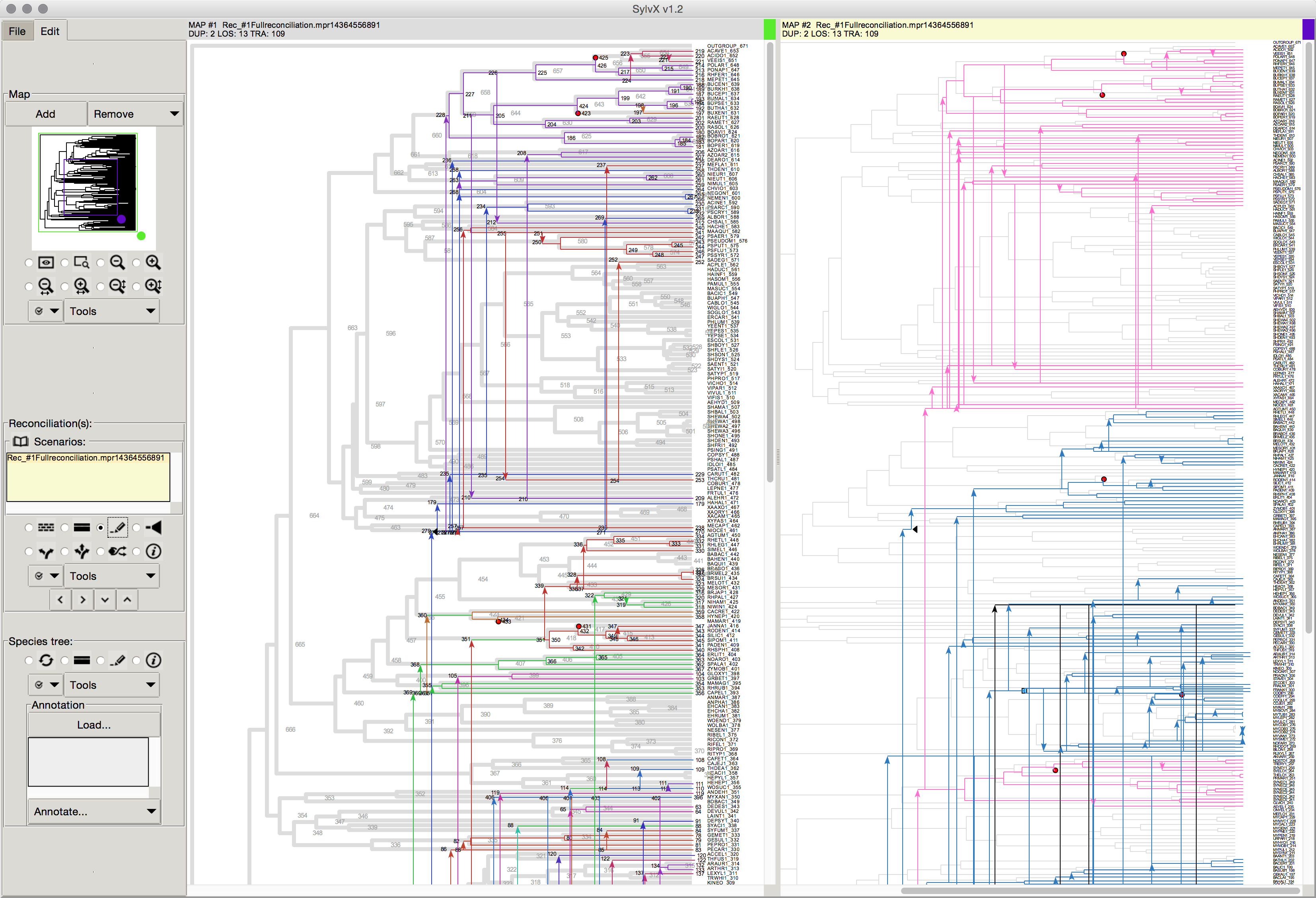
 October 2015 SylvX: a viewer for phylogenetic tree reconciliations Bioinformatics
October 2015 SylvX: a viewer for phylogenetic tree reconciliations Bioinformatics
 March 2015 SylvX: first release
March 2015 SylvX: first release
F. Chevenet
SylvX: a viewer for phylogenetic tree reconciliations
Bioinformatics (2016) 32 (4): 608-610
DOI: https://doi.org/10.1093/bioinformatics/btv625
(*) corresponding author