|
ScripTree: scripting phylogenetic graphics
|
V. 17 IRD-CNRS GEMI-MAB
|
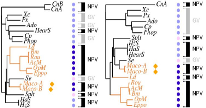
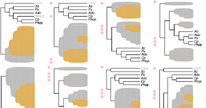
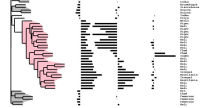
Summary: There is a large amount of tools for interactive display of phylogenetic trees. However, there is a shortage of tools for the automation of tree rendering. Scripting phylogenetic graphics would enable the saving of graphical analyses involving numerous and complex tree handling operations and would allow the automation of repetitive tasks. ScripTree is a tool intended to fill this gap. It is an interpreter to be used in batch mode. Phylogenetic graphics instructions are stored in a text file and performed in a sequential way. Such instructions are related to tree rendering as well as tree annotation. ScriptTree is written in Tcl/Tk making it a cross-platform application, e.g. suitable for Windows and Unix-like systems, including OS X. It can be used either as a standalone package or included in a bioinformatic pipeline and linked to a HTTP server. See the "Examples" section for a ScripTree demonstration by using a web interface. See the "Download & Install" section for downloading ScripTree and its user's manual.
|
This project is supported by grants from:
IRD-SPIRALES 2007-2008
ANR Domaines Emergents 2009-2011 |
ScripTree: scripting phylogenetic graphics
François Chevenet, Olivier Croce, Maxime Hebrard, Richard Christen and Vincent Berry Bioinformatics 2010 26(8):1125-1126 |
|
ScripTree is under the GPL license.
Copyright © 2008 Chevenet F. chevenet@ird.fr/chevenet@lirmm.fr
This program is free software; you can redistribute it and/or
modify it under the terms of the GNU General Public License
as published by the Free Software Foundation; either version 2
of the License, or any later version.
This program is distributed in the hope that it will be useful,
but WITHOUT ANY WARRANTY; without even the implied warranty of
MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. See the
GNU General Public License for more details.
ScripTree source code is here
|