Tutorial : how to create a single phylogenetic tree for bacteria
There are two possibilities to use ScripTree: either using the stand-alone application (you will have to download and install the application onto your system) or directly by using the web version.
The followed examples are made with the online version www.scriptree.org.
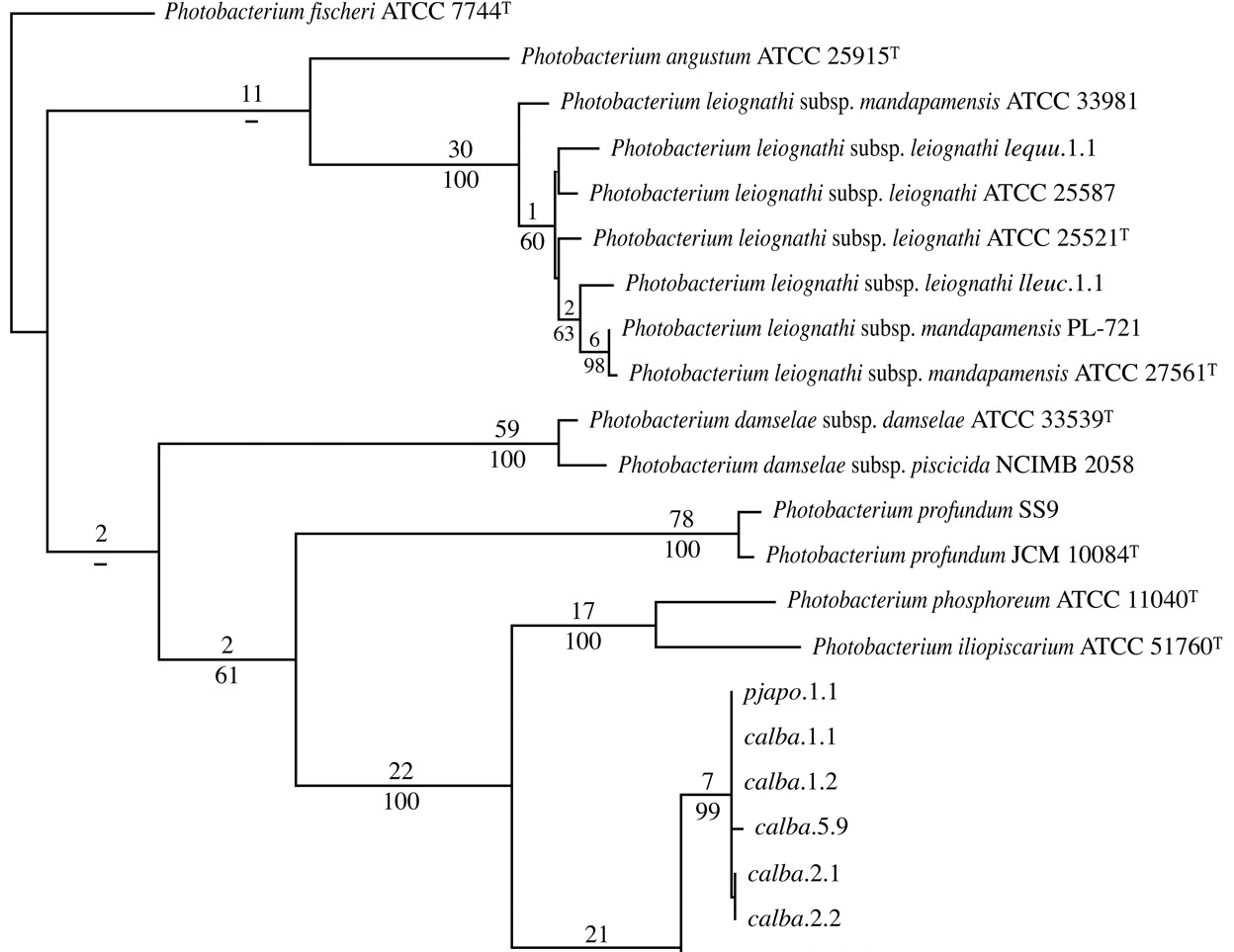
Here, we want to display a tree containing sequences from bacteria. Such trees could be used to highlight a new bacteria specie into a set of well identified bacteria. Moreover, we want to represent this tree in a standard format used in the scientific literature.
It should be mentioned : the names of the genera, the species and the strains, with the letter "T" as exponent if the strain is a « type » strain.
This is an example of a tree for bacteria from a publication [ Paul V. Dunlap1 and Jennifer C., 2005, Applied and Environmental Microbiology]

To build such a figure with ScripTree, we need 3 files:
a file containing the tree into the Newick format;
a file containing the annotations that we want to display for each leaves ;
a file containing the script for Scriptree which describes exactly what should be displayed and how it will be.
* The Newick file
The Newick file will contain the tree that we want to perform. The Newick file should be created from a set of bacterial sequences and by the use of usual tools of phylogeny (Phylip package for example).
Scriptree accepts different variants that may exist for the Newick format (see Scriptree help).
In this example we used a simple format as: ((a, b), c);
The following file contains a Newick tree consisting of sequences of Photobacterium leignathi.
* The annotations file
Annotations are contained in a file different from the Newick file. This file contains various informations that will be displayed for each leaf of the tree. In our example, we want to display the genus and species name, the name of the bacterial strain, a "T" if the strain is a type strain. These informations could be retrieved from the EMBL entries.
These annotations have to be contained in an ascii file which must be in TLF format. This format is a specific format used by the software TreeDyn [Chevenet et al.2006, BMC bioinformatics] (http://www.treedyn.org/).
The TLF file looks like:
|
leaf_name_1 annotation_name_1 {annotation_value_1} {annotation_value_1} ... leaf_name_2 annotation_name_2 {annotation_value_2} {annotation_value_2} … |
Example with real data:
|
AB243239 accession {AB243239} organism {Photobacterium leiognathi} strain {} type {} AB243240 accession {AB243240} organism {Photobacterium leiognathi} strain {} type {} |
AB243239 is the name of a leaf. “accession”, “organism”, “strain” and “type” are the keys that contain (or not) a value or a series of values surrounded by {}.
* The script file
This is the script that will be used (TDS file):
|
tree -linewidth 2 -scaleps 25c -height 1350 l_string_annotation -what organism -how replace -font {Times 12 italic} l_string_annotation -what strain -how juxtapose -font {Times 12 normal} -prefix \ \ \ l_string_annotation -what type -how juxtapose -translate -6 -font {Times 10 normal} |
Explanations:
Line 1: Specifies to display a tree (command "tree") with branches of 2 pixels thick ( "-linewidth 2). "-scaleps 25c" is an option to get an appropriate resolution for the image. "-height 1350" sets the overall height of this tree to 1350 pixels.
Line 2: the command « l_string_annotation » displays the value of the annotation « organism » (« -what organism »). (« -how replace » means that the annotation replaces the default name of the leaf (in our example, the accession number). The option « -font {Times 12 italic} » defines the appearance of the text (italic).
Line 3: Same as the line above, but it displays the annotations "strain" on the right of the previous displayed annotation ( "-how juxtaposes"). The option "-prefix \ \ \" adds 3 blanks before the annotation text (to ensure a good separation with the previous annotations).
Line 4: Shows the value of the annotation "type". If the value is not set (this is not a type strain for example), nothing will be displayed. The value is displayed on the right of the previous one ( "-how juxtapose") and as exponent (little shift to the top with the option "-translate -10").
Now, go to the form of ScripTree online (http://www.scriptree.org) .
There are 3 boxes: "Tree", "Script" and "Annotations". Complete these 3 boxes by pasting the previous informations corresponding to each box (the contents of the files tree.nwk, script.tds and annotations.tlf).
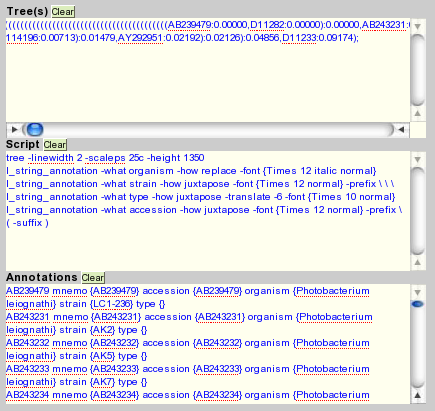
Submit the form ("submit" button). Then, a miniature tree will appear with some links that allow you to download the files containing the tree generated into various formats (the formats are not all available at time of writing this tutorial).

This is the Jpeg file created with the previous data:
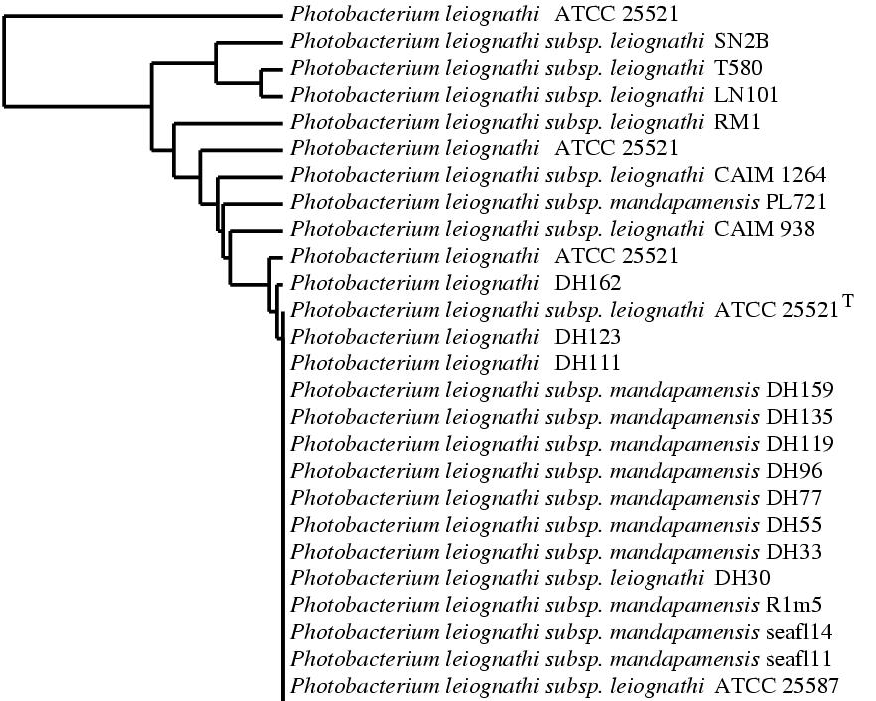
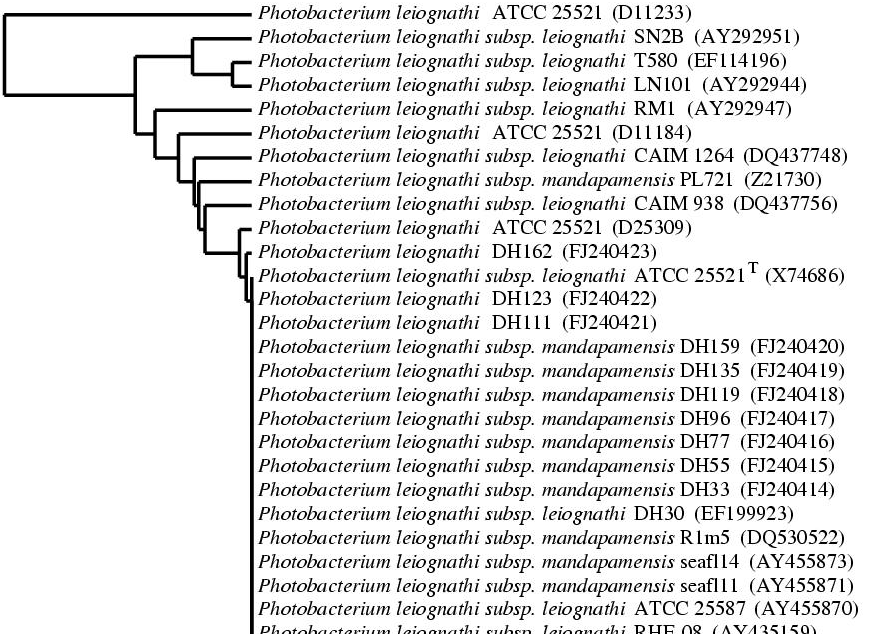
Note that we could add a 5th line to the script in order to see the accession numbers of the sequences.
|
l_string_annotation -what accession -how juxtapose -font {Times 12 normal} -prefix \ ( -suffix ) |
Here, we add the annotation "accession" on the right of the previous annotation (« -how juxtapose »). The option "-prefix \ \ ("-suffix) " allows to add 2 parenthesis for each accession numbers.
The figure becomes :
Of course, we can modify the script to obtain a nicer tree.
In addition, we have to mentioned that a software existed to easily obtain such files used in this example. This software is named “Blast2Tree”. It can automatically create annotations and trees files from a sequences set or from the names of a given micro-organism. For more information, we suggest to have a look on http://bioinfo.unice.fr/blast/ → Blast2Tree.