
- PhyloType Home
|
|
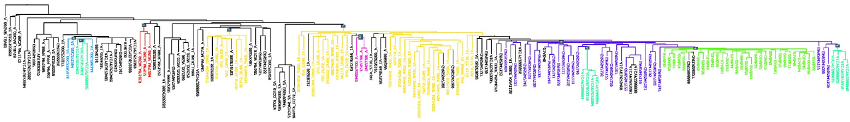
Very large phylogenies are being built today in order to study virus evolution, trace the origin of epidemics, establish the mode of transmission, and survey the appearance of drug resistance. However, no tool is available to quickly inspect these phylogenies and combine them with extrinsic traits (e.g. geographic location, risk group, presence of a given resistance mutation), seeking to extract strain groups of specific interest or requiring surveillance.
|
(see the News section below for more details) July 15 2014. PhyloType batchmodule (PBM), version 07 July 15 2014. PhyloType standalone (PST), version 07 |


|
If you use this site, please cite:
Searching for Virus Phylotypes Chevenet F, Jung M, Peeters M, de Oliveira T, Gascuel O. Bioinformatics (2013) Volume 29, Issue 5Pp. 561-570. Questions and comments much welcome ! chevenet@lirmm.fr, gascuel@lirmm.fr, tdeoliveira@africacentre.ac.za |


|
gascuel@lirmm.fr
tdeoliveira@africacentre.ac.za
- web, pst, pbm : Persistence criteria revisited
- pbm : new parameters file (new output : tree with ancestral annotations and node IDs (in place of support values))




